Guillaume Beslon
My research activity deals with computational biology, artificial life and in silico experimental evolution (aka digital genetics).
I develop and use the aevol model (www.aevol.fr). Aevol is a digital genetics platform specifically designed to study the evolutionary dynamics of microorganisms. The rational of the platform is that the structure of the fitness landscape of an evolving organism is strongly determined by (i) the genotype-to-phenotype map and (ii) the diversity of the mutational operators that affect its genome. Aevol used a fine grained model of both elements, hence enabling to study the molecular evolution.
Ongoing projects:
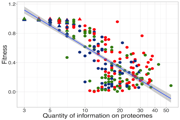
Evolution of complexity: In aevol organisms can evolve different levels of complexity at the genomic, proteomic and phenotypic levels. We use the platform to test the existence of a "complexity ratchet"
Evolution of genome size: We use aevol to investigate the effect of various selective and non-selective forces acting on genome size (both on coding and non-coding compartments).
Prediction of evolution: In aevol mutations occur in a perfect random way but their fixation is likely to be biased by selection. We can use the platform to test whether this enables developping prediction rules.
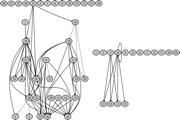
Raevol: Raevol is an extention of aevol developped to study the evolution of genetic regulation networks.
Benchmarking: Using aevol, we are able to produce datasets that can be used to benchmark bioinformatics tools (e.g. phylogenetic software).
Past projects:
Evolution of Evolution: "EvoEvo" was an FP7 EU funded project. The objective of the project was to create an interdisciplinary consortium (experimental evolution, computational biology, artificiel evolution and computer science) to (1) observe how microorganisms uses robustness or evolvability mechanisms to enhance their evolutionary potential (2) model these mechanisms in silico (3) develop an artificial platform able to exploit these mechanisms in software engineering context (4) uses these mechanisms in a real application.
Stochagene: In a long-lasting collaboration with Olivier Gandrillon (ENS-Lyon), we used models to study the origin and consequences of Stochasticity in Gene Expression (SGE). Stochagene was funded by the French National Research Agency (ANR).
Research